Coding Club
We are excited with our ongoing LeedsOmics Coding Club (LOCC) sessions, an open and friendly space where you can bring all your omics analytical issues to discussion on a monthly meeting series. Thanks for all those who have attended and contributed to the club, enriching our chat. It has been great! Please check out the introductory slides we prepared for each session:
Nov/2023-Aug/2024: Dealing with scRNA-seq in R; Dealing with NGS data: Aligning/Mapping reads; Dealing with NGS data: Differential Splicing; Dealing with NGS data: QC and trimming; Dealing with NGS data - DE;
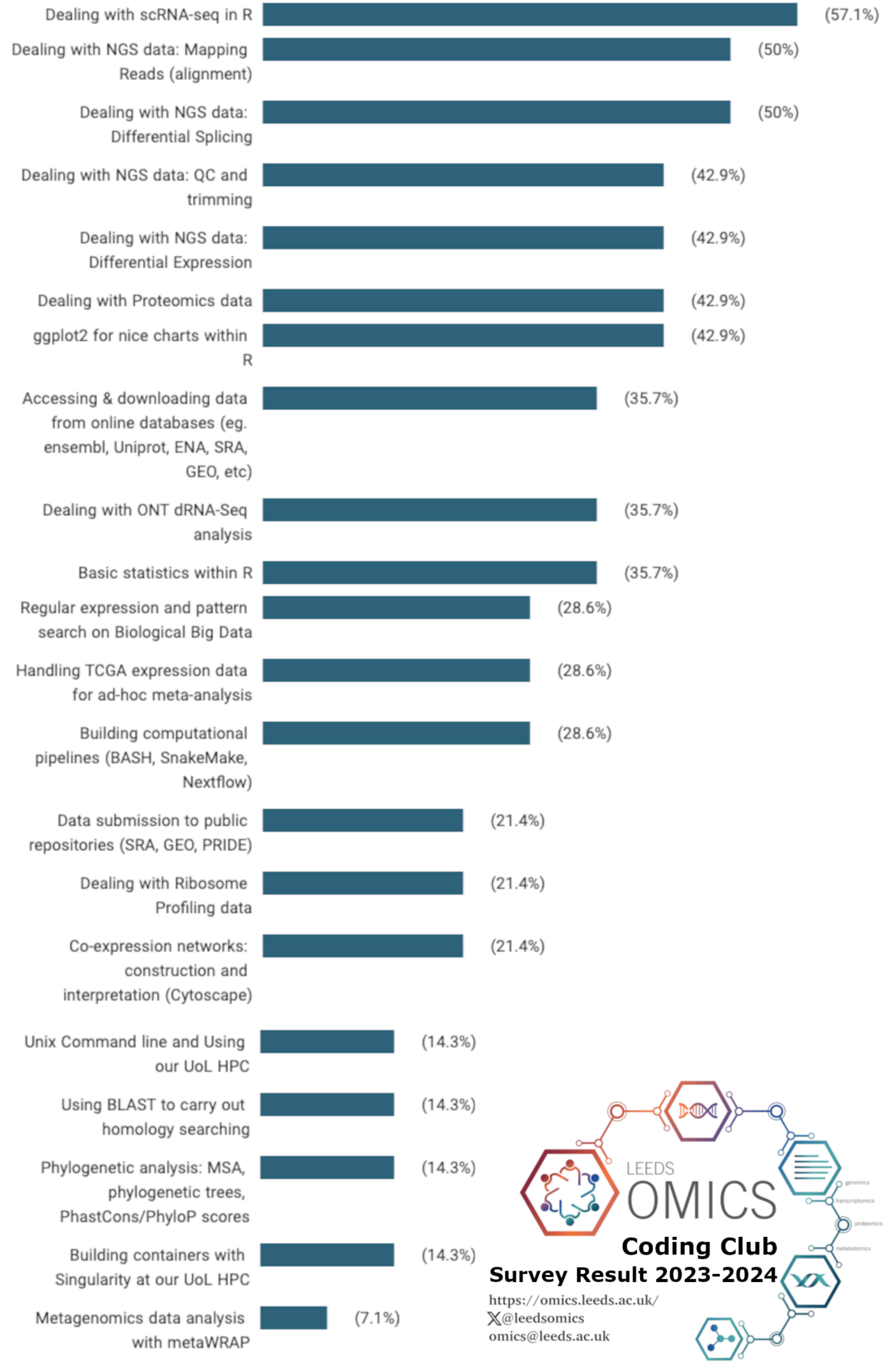
We are organising club sessions by broad themes that LeedsOmics community might be interested in. Therefore, we are continuously launching annual surveys on specific topics which researchers may want to chat on. The chart below depicts the result from the last survey (Oct/2023) sorted by percentage of people interested. As agreed, LOCC sessions' order will follow that result.
Keep an eye on your email inbox in order to receive calendar invites and/or updates on our weekly events through our LeedsOmics mailing list. If you're not subscribed, please do so through this link (you must be logged in at the office365 with your University credentials).
Feel free to drop us an email anytime with more suggestions on topics to be covered (omics@leeds.ac.uk).
Past seasons
Nov/2022-Aug/2023: Dealing with NGS Data: Aligning/Mapping reads; Plotting charts with ggplot2 within R; Accessing & downloading data from online DBs; Basics statistics within R; Dealing with NGS data - DE; Dealing with NGS data - QC and trimming; Dealing with NGS data - Differential Splicing;
Oct/2021-Aug/2022: Good practices in writing code; Basic statistics within R; Plotting charts with ggplot2 within R; Dealing with proteomics data; Unix command line and using HPC at UoLeeds; Accessing & downloading data from online DBs; Dealing with NGS data - QC and trimming; Dealing with NGS data - DE;
Jun/2020-Jul/2021:NGS Quality Control; Accessing & downloading data from online DBs _v2; Good practices in writing code - types of variables; NGS Aligning/Mapping; Good practices in writing code - conditional statements and loopings; NGS - DE; Good practices in writing code - OOP vs Procedural; Plotting charts with ggplot2 within R; Building pipelines with SnakeMake; Basic statistics within R;
Jun/2019-May/2020: ggplot2 for nice charts; Good practice in writing code; Basic statistics within R; Accessing & downloading data from online DBs; Dealing with NGS data; Regular expression and pattern search; Phylogenetics analysis;Using the HPC at UoL; BLAST for sequence similarity search; 16S-based data for Microbiome Analyses;
IMPORTANT NOTE: Please bear in mind that the Coding Club is not supposed to be a tutorial on how to perform bioinformatic analyses from the scratch. You are expected to have already started your analysis, investigated the theme on your own, for then bringing your issues to sessions. Your issues can be either doubts/frustrations during your coding process or comments/hints on best practices in your analyses.
Looking forward to seeing you at the club,
Sincerely,
Elton Vasconcelos (organiser)
