Single Cell Genomics
Single cell genomics is the application of high-throughput approaches to individual cells.
The University of Leeds was awarded £1.1m by the MRC to expand its facilities for single-cell genomics research. The unit is located within the research institutes at St James’s University Hospital and now contains a Fluidigm C1 microfluidics platform and a BD Biosciences FACSeq.
 The C1 currently forms part of Cancer Research UK Leeds Centre Genomics facility. It enables capture of up to 96 single cells on a microfluidic chip, followed by washing and lysis of the cells and subsequent cDNA amplification and analysis. The latter depends upon the researcher’s requirements and can either consist of:
The C1 currently forms part of Cancer Research UK Leeds Centre Genomics facility. It enables capture of up to 96 single cells on a microfluidic chip, followed by washing and lysis of the cells and subsequent cDNA amplification and analysis. The latter depends upon the researcher’s requirements and can either consist of:
- Targeted amplification of up to 96 genes with subsequent analysis on Fluidigm’s Biomark system enabling qPCR of all 96 genes in the 96 isolated single cells
- Whole transcriptome capture within each cell and subsequent mRNAseq
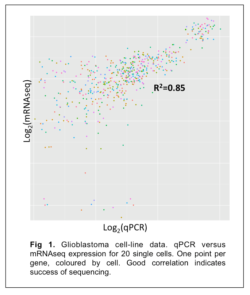
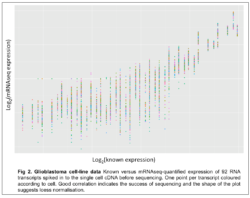
When performing the latter, it is also possible to use some of the amplified cDNA for validation via the qPCR route. This pipeline has been established using brain cancer (Glioblastoma) cell lines with subsequent comparison of the expression by mRNAseq with A) qPCR of genes in the same cells (Fig.1), and B) the known input concentration of ERCC spike in RNAs (Fig.2) both indicating the accuracy of the method.
 There are continual developments of this platform yielding more sensitive and economical workflows such as the Smart-seq2 protocol now being tested for adoption in Leeds (1,2).
There are continual developments of this platform yielding more sensitive and economical workflows such as the Smart-seq2 protocol now being tested for adoption in Leeds (1,2).
The FACSeq is a desktop FACS machine designed specifically for sorting single cells into 96- or 384-well plates in under a minute. This machine can be run with minimal training and will facilitate single cell processing of numerous tissues collected by the biobanking facility in Leeds. The manufacturers of the machine, BD Biosciences, offer protocols for targeted or whole transcriptome amplification of cDNA within the sorted cells, with unique molecular indexing to account for the biases that can be introduced with PCR.
In LeedsOmics, the Stead Group use single cell genomics approaches to understand intratumour heterogeneity in malignant brain cancer and how this may be responsible for inevitable tumour regrowth following current therapy regimens (Yorkshire Cancer Research, Brain Tumour Research and Support, and Ellie’s Fund grant). Read more about the glioma group at Leeds here.
References:
1. Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protocols. 2014 01//print;9(1):171-81.
2. Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Meth. 2013 11//print;10(11):1096-8.
