Translatomics
Translatomics is the study of the translational status of the entire transcriptome.
There are many steps involved that contribute to gene expression. Many people consider mRNA levels (RNA-seq) to be the main read out of gene expression. However, it is just one of many indicators. Gene expression can be highly regulated at the point of mRNA translation, especially during translation initiation.
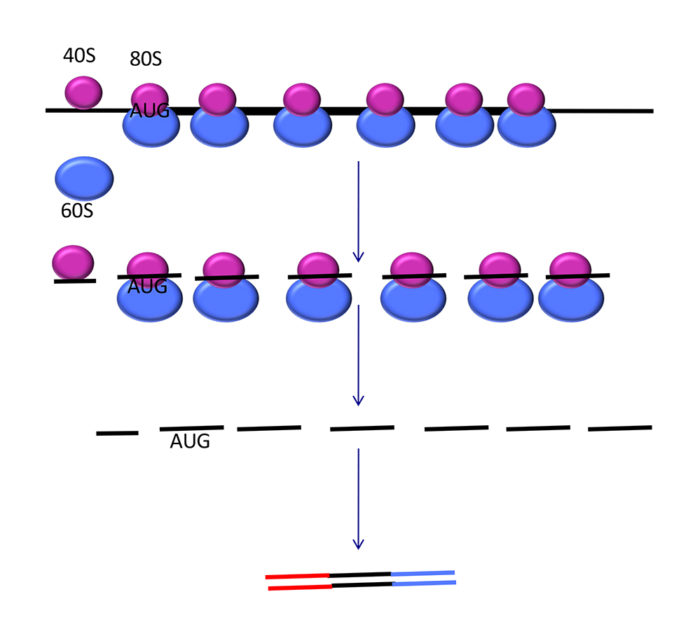
The primary technique used to study translatomics is ribosome profiling (Ribo-Seq), which was developed in 2009 by Nick Ingolia (UC Berkeley) and Jonathan Weismann (UCSF). This approach has revolutionised the way we study translation. Since then there have been many adaptations and improvements both experimentally and computationally to understand the ribosome footprints that are sequenced.
During ribosome profiling the exact position of all ribosomes across the transcriptome is determined. RNases are used to degrade RNA that is not protected by bound ribosomes. These protected RNA fragments, ribosome footprints are 28-32nt in length depending on the RNase used or the organism profiled. These footprints are purified and subject to Next Generation Sequencing along with the total poly-adenylated RNA as a control. As a result of the way ribosomes move along mRNAs decoding codons in triplets, the frame of translation can be detected in ribosome profiling data. There is a bias of the RNA reads to the codon frame, mapping the 5’ end of reads reveals the frame because the majority of reads result from the ribosome decoding at the P site. Start codons can also be identified through footprint accumulation and the use of drugs that stall ribosomes at the start codon. Therefore, Ribo-Seq has helped reveal new points of initiation, new open reading frames, new points of termination and new frames of translation.
In LeedsOmics, the Aspden Group use Ribo-Seq to identify novel regions of translational in neurons (MRC New Investigator Research Grant) and to understand how translation is regulated during Drosophila spermatogenesis (BBSRC PhD studentship). @RNA_Julie / http://www.fbs.leeds.ac.uk/staff/profile.php?tag=Aspden_J